- 532.27 KB
- 2022-04-22 13:43:24 发布
- 1、本文档共5页,可阅读全部内容。
- 2、本文档内容版权归属内容提供方,所产生的收益全部归内容提供方所有。如果您对本文有版权争议,可选择认领,认领后既往收益都归您。
- 3、本文档由用户上传,本站不保证质量和数量令人满意,可能有诸多瑕疵,付费之前,请仔细先通过免费阅读内容等途径辨别内容交易风险。如存在严重挂羊头卖狗肉之情形,可联系本站下载客服投诉处理。
- 文档侵权举报电话:19940600175。
'中国科技论文在线http://www.paper.edu.cnMitochondrialgenomesequencingofthreefireflybeetles#andphylogeneticanalysisof18speciesofbeetles**5WANGKai,HONGWei,JIAOHengwu,MUFeng-Juan,ZHAOHuabin(CollegeofLifeSciences,WuhanUniversity,Wuhan430072)Abstract:Tovalidatethephylogeneticinferencebasedontranscriptomedata,weemployedthetraditionalSangersequencingtoobtaintwocompletemitogenomes(AquaticafictaandA.wuhana)andonenearlycompletemitogenome(Lamprigerayunnana);wealsoidentifiedthe13proteincodinggenes10(PCGs)inmitogenomesfromeachofthefourwhole-bodytranscriptomes;andweadditionallyretrieved12publishedcompletemitogenomesfrommembersofRhagophthalmidae,Lampyridae,Phengodidae,Elateridae,Lycidae,Cantharidae,andTenebrionidae.OurmitogenomeanalysisconfirmedthedistinctivenessofRhagophthalmidaefromLampyridae,whichisconsistenttoourearliertranscriptomeanalysis.15Keywords:Lampyridae,Phylogenomics,Mitochondrialgenome,Mitogenome0IntroductionTheoriginandevolutionofbioluminescenceinbeetleshaspromptedseveralstudiestoilluminate[1-8]thephylogeneticrelationshipswithinbioluminescentbeetles.However,thephylogenetic20positionofRhagophthalmidaehasbeenalong-standingpuzzle.Therearethreemajorhypothesesconcerningthisissue.First,RhagophthalmidaehadbeenassignedtothefamilyPhengodidae[3]inferredfrommorphologicaldata.Second,RhagophthalmidaehadbeenconsideredtobeasubfamilyorgenusinthefamilyLampyridaebasedonmorphological,embryologicaland[4,6,7,9]molecularevidence.Third,morerecentevidencefrombothmorphologicalandmolecular25datahassupportedthefamilialstatusofRhagophthalmidae,whichisdistinctfromboth[1,2,5,8]LampyridaeandPhengodidae.Giventhattheevolutionaryhistoryofmorphologicalfeaturesisusuallycomplex,molecularevidencehasbeenexpectedtoresolvetaxonomicstatusand[10]phylogeneticrelationshipsthathavebeencontentiousbasedonmorphologicalevidence.However,theincongruenceofmolecularevidencehasbeenobservedinphylogeneticanalyses[1,5,7]30concerningthephylogeneticpositionofRhagophthalmidae,possiblybecauseasingleorafew[11]geneslacksufficientphylogeneticsignals.Inarecentstudy,weemployedtheIlluminaHiseq2000platformandsequencedthewhole-bodytranscriptomesofthefourluminescentbeetles:onerhagophthalmidbeetle(Rhagophthalmussp.)35andthreerepresentativesoflampyridbeetles(Asymmetricatacircumdata,Aquaticaficta,andPyrocoeliapectoralis).PhylogenomicanalysisusingthesetranscriptomedataappearedtorecognizethedistinctivenessofRhagophthalmidaefromLampyridae.Tovalidatethephylogeneticinferencebasedontranscriptomedata,weemployedthetraditionalSangersequencingtoobtaintwocompletemitogenomes(AquaticafictaandA.wuhana)andonenearlycompletemitogenome40(Lamprigerayunnana);wealsoidentifiedthe13proteincodinggenes(PCGs)inmitogenomesfromeachofthefourwhole-bodytranscriptomes(Table1);andweadditionallyretrieved12publishedcompletemitogenomesfrommembersofRhagophthalmidae,Lampyridae,Phengodidae,Elateridae,Lycidae,Cantharidae,andTenebrionidae(seethespeciesnamesandGenBankaccessionsinMaterialsandMethods).Ourmitogenomeanalysisconfirmedthedistinctivenessof45RhagophthalmidaefromLampyridae.Foundations:TheSpecializedResearchFundfortheDoctoralProgramofHigherEducation(20130141110066).Briefauthorintroduction:WANGKai(1986-),Male,PhDcandidate,AnimalEvolutionCorrespondanceauthor:ZHAOHuabin(1980-),Male,Professor,AnimalEvolutionandEcologicalGenomics.E-mail:huabinzhao@whu.edu.cn-1-
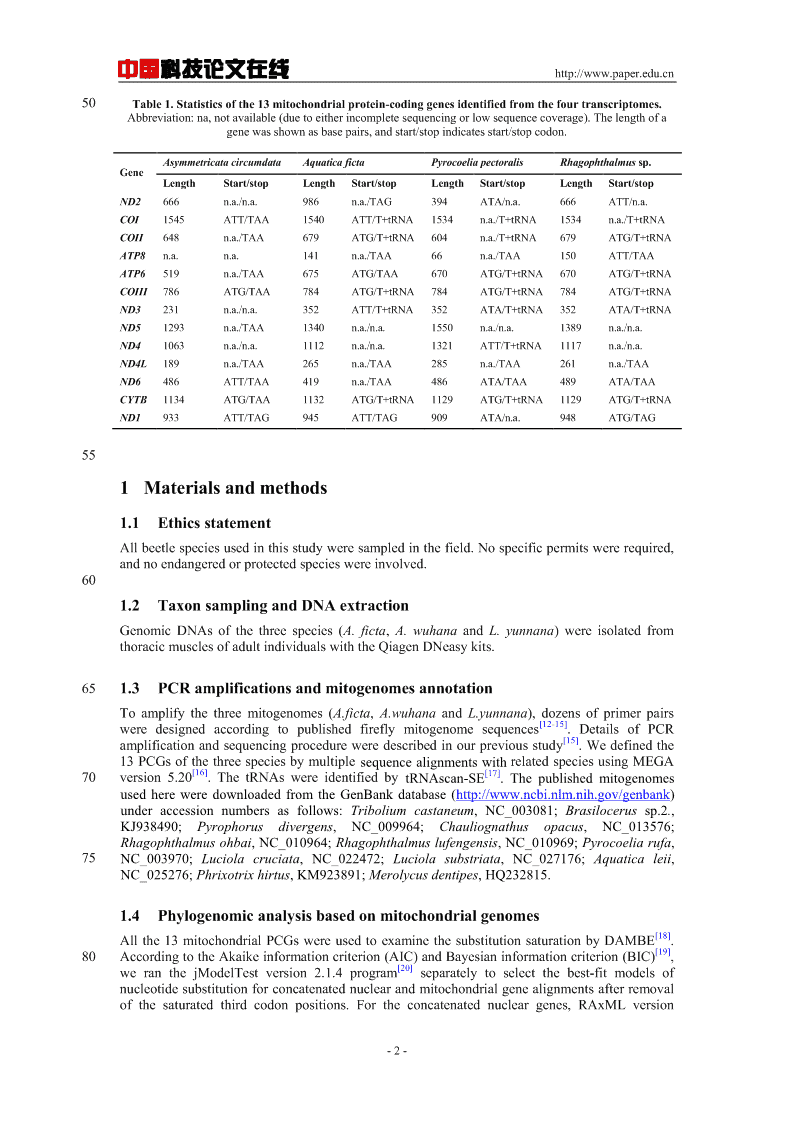
中国科技论文在线http://www.paper.edu.cn50Table1.Statisticsofthe13mitochondrialprotein-codinggenesidentifiedfromthefourtranscriptomes.Abbreviation:na,notavailable(duetoeitherincompletesequencingorlowsequencecoverage).Thelengthofagenewasshownasbasepairs,andstart/stopindicatesstart/stopcodon.AsymmetricatacircumdataAquaticafictaPyrocoeliapectoralisRhagophthalmussp.GeneLengthStart/stopLengthStart/stopLengthStart/stopLengthStart/stopND2666n.a./n.a.986n.a./TAG394ATA/n.a.666ATT/n.a.COI1545ATT/TAA1540ATT/T+tRNA1534n.a./T+tRNA1534n.a./T+tRNACOII648n.a./TAA679ATG/T+tRNA604n.a./T+tRNA679ATG/T+tRNAATP8n.a.n.a.141n.a./TAA66n.a./TAA150ATT/TAAATP6519n.a./TAA675ATG/TAA670ATG/T+tRNA670ATG/T+tRNACOIII786ATG/TAA784ATG/T+tRNA784ATG/T+tRNA784ATG/T+tRNAND3231n.a./n.a.352ATT/T+tRNA352ATA/T+tRNA352ATA/T+tRNAND51293n.a./TAA1340n.a./n.a.1550n.a./n.a.1389n.a./n.a.ND41063n.a./n.a.1112n.a./n.a.1321ATT/T+tRNA1117n.a./n.a.ND4L189n.a./TAA265n.a./TAA285n.a./TAA261n.a./TAAND6486ATT/TAA419n.a./TAA486ATA/TAA489ATA/TAACYTB1134ATG/TAA1132ATG/T+tRNA1129ATG/T+tRNA1129ATG/T+tRNAND1933ATT/TAG945ATT/TAG909ATA/n.a.948ATG/TAG551Materialsandmethods1.1EthicsstatementAllbeetlespeciesusedinthisstudyweresampledinthefield.Nospecificpermitswererequired,andnoendangeredorprotectedspecieswereinvolved.601.2TaxonsamplingandDNAextractionGenomicDNAsofthethreespecies(A.ficta,A.wuhanaandL.yunnana)wereisolatedfromthoracicmusclesofadultindividualswiththeQiagenDNeasykits.651.3PCRamplificationsandmitogenomesannotationToamplifythethreemitogenomes(A.ficta,A.wuhanaandL.yunnana),dozensofprimerpairs[12-15]weredesignedaccordingtopublishedfireflymitogenomesequences.DetailsofPCR[15]amplificationandsequencingprocedureweredescribedinourpreviousstudy.Wedefinedthe13PCGsofthethreespeciesbymultiplesequencealignmentswithrelatedspeciesusingMEGA[16][17]70version5.20.ThetRNAswereidentifiedbytRNAscan-SE.ThepublishedmitogenomesusedhereweredownloadedfromtheGenBankdatabase(http://www.ncbi.nlm.nih.gov/genbank)underaccessionnumbersasfollows:Triboliumcastaneum,NC_003081;Brasilocerussp.2.,KJ938490;Pyrophorusdivergens,NC_009964;Chauliognathusopacus,NC_013576;Rhagophthalmusohbai,NC_010964;Rhagophthalmuslufengensis,NC_010969;Pyrocoeliarufa,75NC_003970;Luciolacruciata,NC_022472;Luciolasubstriata,NC_027176;Aquaticaleii,NC_025276;Phrixotrixhirtus,KM923891;Merolycusdentipes,HQ232815.1.4Phylogenomicanalysisbasedonmitochondrialgenomes[18]Allthe13mitochondrialPCGswereusedtoexaminethesubstitutionsaturationbyDAMBE.[19]80AccordingtotheAkaikeinformationcriterion(AIC)andBayesianinformationcriterion(BIC),[20]weranthejModelTestversion2.1.4programseparatelytoselectthebest-fitmodelsofnucleotidesubstitutionforconcatenatednuclearandmitochondrialgenealignmentsafterremovalofthesaturatedthirdcodonpositions.Fortheconcatenatednucleargenes,RAxMLversion-2-
中国科技论文在线http://www.paper.edu.cn[21]7.2.6wasusedtoreconstructtheMLtreeundertheGTR+GAMMAmodelwith100bootstrap[22]85replicates,andMrBayesversion3.2.6wasappliedtoreconstructtheBItreeusingthe[23,24]recommendedGTR+I+Gmodelwith0.5milliongenerations.Toreducetheimpactofgenetreeheterogeneity,wealsoundertookphylogeneticanalysiswiththecoalescentmodelusingthe[25]MP-ESTmethod.Fortheconcatenated13mitochondrialPCGs,theMLtreewasreconstructedbyRAxMLversion7.2.6with1000bootstrapreplicatesunderrecommendedGTR+GAMMA90model,andtheconcatenatedsequencewaspartitionedbydifferentgenestoestimateandoptimizeindividualα-shapeparameters,GTR-rates,andbasefrequenciesforeachgene.MrBayesversion3.2.6wasusedtoreconstructtheBItreewithonemilliongenerationsandtheconcatenatedsequencewaspartitionedaccordingtodifferentmodels:HKY+GmodelforND4L,GTR+GmodelforND1andATP6,andGTR+I+Gmodelfortheother10mitochondrialgenes.95Inaddition,weuseddeducedproteinsequencesinourphylogenomicanalysis,aimingtoreducetheimpactofnucleotidecompositionalbias.Briefly,proteinsequencesofmitochondrialgenes[16][26]werededucedandalignedbyMEGAversion5.20.ProtTestversion3.41wasappliedtoselectthebest-fitmodelofproteinsequenceevolutionfollowingtheAkaikeinformationcriterion100(AIC).TheMtRev+I+Gmodelwasselectedfortheconcatenatedproteinsequencesof[22]mitochondrialgenes.PhylogenetictreeswerereconstructedbyMrBayesversion3.2.6withasmanygenerationsasrequired.2ResultsandDiscussion1052.1MitogenomesequencingUsingthetraditionalSangersequencingapproach,wesequencedtwocompletemitogenomes(A.fictaandA.wuhana)andonenearlycompletemitogenome(L.yunnana)(Table2).WewereunabletosequencetheregionbetweenND2and12SrRNAinL.yunnanadespitemultipleattempts(Table2),possiblybecausethisregioncontainstheA+T-richregionwhichmaypose[27]110technicalissuesinsequencing.Allthe13proteincodinggenes(PCGs)fromeachofthethreemitogenomeswerecompleteexcepttheND2inL.yunnanalackingasegmentnearits5’end(Table2).Allthe13PCGs,2rRNAs,22transferRNAs(tRNAs)and1A-T-richregioncommon[28]tothevastmajorityofanimalmitogenomeswereidentifiedinthethreenewlysequencedTrpCysTyrmitogenomes,exceptforthethreetRNAs(tRNA,tRNA,tRNA)andtheA-T-richregionin115L.yunnana(Table2).Arrangementsandorientationsofallgenesinthethreemitogenomesare[12-15]identicaltootherbeetles.Similartootherfireflybeetles,allthePCGsemployedtraditionalmitochondrialstartcodonsATNandterminatedwithTAA,TAGorsingleT(Table2).2.2Phylogeneticanalysisbasedonmitochondrialgenesequences120Phylogeneticanalysiswasalsoconductedwithmitochondrialgenesequences.Firstly,weretrievedpublishedmitogenomesfrom12speciesofrelatedbeetles:Rhagophthalmusohbai(Rhagophthalmidae),Rhagophthalmuslufengensis(Rhagophthalmidae),Pyrocoeliarufa(Lampyridae),Luciolacruciate(Lampyridae),Luciolasubstriata(Lampyridae),Aquaticaleii(Lampyridae),Brasilocerussp.2(Phengodidae),Phrixotrixhirtus(Phengodidae),Pyrophorus125divergens(Elateridae),Merolycusdentipes(Lycidae),Chauliognathusopacus(Cantharidae),andTriboliumcastaneum(Tenebrionidae)(Fig.1).Secondly,wenewlydeterminedmitogenomeswiththetraditionalSangersequencingfromthreefireflybeetles:A.ficta(Lampyridae),A.wuhana(Lampyridae)andL.yunnana(Lampyridae)(Table2).Thirdly,weidentifiedmitochondrialgenesequencesfromournewlyderterminedtranscriptomeassembliesoffourbeetles:Rhagophthalmus130sp.(Rhagophthalmidae),A.ficta(Lampyridae),A.circumdata(Lampyridae),andP.pectoralis(Lampyridae)(Table1).ThefireflyA.fictahassequencedatafrombothSangerandIlluminasequencing,butthetranscriptome-derivedmitogenomesequenceisincomplete(Table1),wethusselectedtheSanger-basedmitogenomesequence(Table2)forfurtheranalysis.Intotal,ourdatasetofmitogenomescontained18beetles.InadditiontotheoutgroupspeciesT.castaneum,six135familiesofbeetleswereincluded:threerhagophthalmids,ninelampyrids,twophengodids,oneelaterid,onelycid,andonecantharid(Fig.1).-3-
中国科技论文在线http://www.paper.edu.cnTable2.Annotationsofthethreenewlysequencedmitochondrialgenomes.Incompletesequenced140regionwasindicatedwithanasterisk.Abbreviation:n.a.,notavailable(duetoincompletesequencing);Dir.,direction;F,forward;R,reverse;Start/stopindicatesstart/stopcodon.AquaticawuhanaAquaticafictaLamprigerayunnanaGeneDir.FromToStart/stopFromToStart/stopFromToStart/stopIletRNAF164163GlntRNAF6213061129MettRNAR130195129194*ND2F1961209ATA/TAA1951208ATA/TAG1878n.a./TAATrptRNAF1211127512101274882942CystRNAR134214041340140211431203TyrtRNAR140414661402146512031265COIF14383003ATT/TAA14583002ATT/TAA12372802ATT/TAALeutRNAF299930622988306127982861COIIF30643742ATG/T+tRNA30633741ATG/T+tRNA28353540ATA/T+tRNALystRNAF374338133742381235413610AsptRNAF381338763812387436103675ATP8F38774032ATT/TAA38754030ATT/TAA36853828ATA/TAAATP6F40264700ATG/TAA40244698ATG/TAA38224485ATG/T+tRNACOIIIF47005483ATG/T+tRNA46985481ATG/T+tRNA44865266ATG/T+tRNAGlytRNAF548455465482554552675328ND3F55475900ATA/TAG55465899ATT/TAG53355682ATA/TAGAlatRNAF589959625898596156815744ArgtRNAF596260265961602557435805AsntRNAF602660906025609058045865SertRNAF609161576091615758585920GlutRNAF615862206158622159215983PhetRNAR621962816220628359826041ND5R62827989ATT/T+tRNA62847994ATA/T+tRNA60397749ATT/T+tRNAHistRNAR799080527992805477507810ND4R80539379ATG/T+tRNA80559381ATG/T+tRNA78109135ATG/TAGND4LR93739663ATG/TAA93759665ATG/TAA91299416ATG/TAAThrtRNAF966597279667972894189480ProtRNAR972797909729979494819544ND6F979210277ATA/TAA979610284ATA/TAA954610049ATT/TAACYTBF1027711410ATG/TAG1028411417ATG/TAG1005011177ATG/TAGSertRNAF114091147411416114811117611240ND1R1147912437ATT/TAG1149912443ATT/TAG1125712207ATG/TAGLeutRNAR12445125061245112513122091226916SrRNAR125071377312514137761227013525ValtRNAR137741384213777138461352513592*12SrRNAR138431458813847145961358814138A+T-rich14589161861459716836n.a.region145The13mitochondrialPCGsfromeachofthe18beetlespecieswerealignedandconcatenated.SubstitutionsaturationanalysisrevealedthatthethirdcodonsitesofallexaminedmitochondrialPCGshaveexperiencedsubstantialsubstitutionsaturation,whilethefirstandsecondcodonsiteshavenot(Table3),wethusjustusedthefirstandsecondcodonsitesofthe13mitochondrialgenesforsubsequentphylogeneticanalysiswiththeconcatenationmethod.Theconcatenated-4-
中国科技论文在线http://www.paper.edu.cn150alignmentofthe13mitochondrialPCGsis7382ntinlength.BothMLandBIphylogeneticanalysesrecoveredidenticalphylogenetictrees(Fig.1).OurtreesshowedthatRhagophthalmussp.clusteredwithtwootherrhagophthalmidswithpublishedmitogenomes,andthethreerhagophthalmidsformedamonophyleticgroupwithastrongsupportof100%inbothMLandBIanalyses(Fig.1),confirmingthatoursampledRhagophthalmusbeetleindeedbelongsto155Rhagophthalmidaephylogenetically.Asdepictedfromthetrees,RhagophthalmidaewasfirstalliedwithPhengodidaeassistergroups,andRhagophthalmidaeandPhengodidaeformedamonophyleticgroupwithLampyridae(Fig.1),confirmingthatRhagophthalmidaeisnotasubgroupwithinLampyridae.ThelongbranchesinPhengodidaespecieswouldnotaffectourphylogeneticanalysis,becausewerecoveredsimilartreesafterremovingthetwophengodids(Fig.1602).Wedidnotperformthecoalescentmethodinthephylogeneticanalysisbasedonmitogenomes,becausemitochondrialgenesexhibitlimitedincongruenceandtheimpactofmitochondrialgene[29]treeheterogeneityhasbeenbelievedtobeminimal.Aquaticaleii100/100100/100Aquaticaficta100/100Aquaticawuhana100/100LuciolinaeLuciolacruciata100/100LuciolasubstriataLampyridaeAsymmetricatacircumdata100/100Pyrocoeliarufa100/100PyrocoeliapectoralisLampyrinae73/100Lamprigerayunnana77/100Rhagophthalmusohbai100/100100/100RhagophthalmuslufengensisRhagophthalmidaeRhagophthalmussp.50/9981/100Brasilocerussp.2100/100PhengodidaePhrixotrixhirtusElateroidea100/100PyrophorusdivergensElateridae100/100MerolycusdentipesLycidaeChauliognathusopacusCantharidaeTriboliumcastaneumTenebrionidae0.07165Figure1.PhylogeneticrelationshipsbetweenRhagophthalmidaeanditsrelatedbeetlefamiliesinferredfromtheconcatenated13mitochondrialproteincodinggeneswiththethirdcodonpositionsremoved.NumbersatnodesaretheMLbootstrapvalues/Bayesianposteriorprobabilities,shownaspercentages.SpeciesshowninbluehavemitochondrialgenesgeneratedfromSanger170sequencing,whilespeciesinredhavemitochondrialgenesidentifiedfromIllumina-sequencedtranscriptomes.3ConclusionInthispaper,weemployedthetraditionalSangersequencingtoobtaintwocompletemitogenomes175(AquaticafictaandA.wuhana)andonenearlycompletemitogenome(Lamprigerayunnana);wealsoidentifiedthe13proteincodinggenesinmitogenomesfromeachofthefourwhole-bodytranscriptomes;andweadditionallyretrieved12publishedcompletemitogenomesfrommembersofRhagophthalmidae,Lampyridae,Phengodidae,Elateridae,Lycidae,Cantharidae,andTenebrionidae.OurmitogenomeanalysisshowedthatRhagophthalmidaewasfirstalliedwith-5-
中国科技论文在线http://www.paper.edu.cn180Phengodidaeassistergroups,andthatRhagophthalmidaeandPhengodidaeformedamonophyleticgroupwithLampyridae(Fig.1),confirmingthatRhagophthalmidaeisnotasubgroupwithinLampyridae.Aquaticaleii100/100100/100Aquaticaficta100/100Aquaticawuhana100/100Luciolacruciata100/100LuciolasubstriataLampyridaeAsymmetricatacircumdata99/100Pyrocoeliarufa100/100Pyrocoeliapectoralis66/10088/100LamprigerayunnanaRhagophthalmusohbai100/10052/99100/100RhagophthalmuslufengensisRhagophthalmidaeRhagophthalmussp.100/100PyrophorusdivergensElateridae100/100MerolycusdentipesLycidaeChauliognathusopacusCantharidaeTriboliumcastaneumTenebrionidae0.06Figure2.Phylogenetictreesreconstructedfromtheconcatenated13mitochondrialprotein185codinggenes,afterremovingthetwophengodids.NumbersatnodesaretheMLbootstrapvalues/Bayesianposteriorprobabilities,shownaspercentages.Table3.SubstitutionsaturationanalysisofmitochondrialgenesbyDAMBE.190Abbreviations:N,no;Y,yes.DAMBEresultforthe3rdsitesSubstitutionsaturationofSubstitutionsaturationofGenestthe1and2ndcodonsitesthe3rdcodonsiteIssIss.c(S)ProbabilityATP6N0.77290.60870.0000YCOX1N0.68690.71430.1490YCOX2N0.74360.62590.0001YCOX3N0.70530.64820.0351YCYTBN0.70600.68380.2703YND1N0.70310.66460.1471YND2N0.71540.50350.0001YND3N0.75260.47800.0000YND4N0.74230.67950.0110YND4LN0.73050.50290.0002YND5N0.73250.69150.0655YND6N0.79180.57140.0000Y-6-
中国科技论文在线http://www.paper.edu.cnAcknowledgements(Optional)195WethankDr.XinhuaFu(HuazhongAgriculturalUniversity)whokindlyprovidedthetwospeciesoffireflybeetlesamples.WethankDr.ChengquanCao(LeshanNormalUniversity)forcollectingthesampleofLamprigerayunnana.WealsothankYiWangandLiangAo(WuhanUniversity)fortechnicalassistanceinthelaboratory.ThisworkwassupportedinpartbytheSpecializedResearchFundfortheDoctoralProgramofHigherEducation(20130141110066).200References1Amaral,D.T.,Mitani,Y.,Ohmiya,Y.&Viviani,V.R.OrganizationandcomparativeanalysisofthemitochondrialgenomesofbioluminescentElateroidea(Coleoptera:Polyphaga).Gene586,254-262(2016).2052Branham,M.A.&Wenzel,J.W.Theevolutionofbioluminescenceincantharoids(Coleoptera:Elateroidea).Fla.Entomol.84,565-586(2001).3Crowson,R.A.AreviewoftheclassificationofCantharoidea(Coleoptera)withthedefinitionoftwonewfamilies,CneoglossidaeandOmethidae.Rev.Univ.Madrid21,35–77(1972).4Kobayashi,Y.,Suzuiki,H.&Ohba,N.Formationofasphericalgermrudimentintheglow-worm210RhagophthalmusohbaiWittmer(Coleoptera:Rhagophthalmidae)anditsphylogeneticimplications.Proc.Arthropod.Embryol.Soc.Jpn.36,1-5(2001).5Kundrata,R.,Bocakova,M.&Bocak,L.ThecomprehensivephylogenyofthesuperfamilyElateroidea(Coleoptera:Elateriformia).Mol.Phylogenet.Evol.76,162-171(2014).6McDermott,F.A.ThetaxonomyoftheLampyridae(Coleoptera).Trans.Am.Entomol.Soc.90,1-72215(1964).7Stanger-Hall,K.F.,Lloyd,J.E.&Hillis,D.M.PhylogenyofNorthAmericanfireflies(Coleoptera:Lampyridae):Implicationsfortheevolutionoflightsignals.MolPhylogenetEvol45,33-49(2007).8Wittmer,W.&Ohba,N.NeueRhagophthalmidae(Coleoptera)ausChinaundbenachbartenLändern.Jpn.Entomol.62,341–355(1994).2209Li,X.Y.,Yang,S.,Xie,M.&Liang,X.C.Phylogenyoffireflies(Coleoptera:Lampyridae)inferredfrommitochondrial16SribosomalDNA,withreferencestomorphologicalandethologicaltraits.ProgNatSci16,817-826(2006).10Nei,M.&Kumar,S.MolecularEvolutionandPhylogenetics.(OxfordUniversityPress,2000).11Jeffroy,O.,Brinkmann,H.,Delsuc,F.&Philippe,H.Phylogenomics:thebeginningofincongruence?225TrendsGenet22,225-231(2006).12Bae,J.S.,Kim,I.,Sohn,H.D.&Jin,B.R.Themitochondrialgenomeofthefirefly,Pyrocoeliarufa:completeDNAsequence,genomeorganization,andphylogeneticanalysiswithotherinsects.Mol.Phylogenet.Evol.32,978-985(2004).13Li,X.,Ogoh,K.,Ohba,N.,Liang,X.C.&Ohmiya,Y.Mitochondrialgenomesoftwoluminousbeetles,230RhagophthalmuslufengensisandR-ohbai(Arthropoda,Insecta,Coleoptera).Gene392,196-205(2007).14Mu,F.J.,Ao,L.,Zhao,H.B.&Wang,K.Characterizationofthecompletemitochondrialgenomeofthefirefly,Luciolasubstriata(Coleoptera:Lampyridae).MitochondrialDNA,1-3(2015).15Jiao,H.,Ding,M.&Zhao,H.Sequenceandorganizationofcompletemitochondrialgenomeofthefirefly,Aquaticaleii(Coleoptera:Lampyridae).MitochondrialDNA.0,1-2(2013).23516Tamura,K.etal.MEGA5:molecularevolutionarygeneticsanalysisusingmaximumlikelihood,evolutionarydistance,andmaximumparsimonymethods.Mol.Biol.Evol.28,2731-2739(2011).17Lowe,T.M.&Eddy,S.R.tRNAscan-SE:aprogramforimproveddetectionoftransferRNAgenesingenomicsequence.NucleicAcidsRes.25,955-964(1997).18Xia,X.H.DAMBE5:Acomprehensivesoftwarepackagefordataanalysisinmolecularbiologyand240evolution.Mol.Biol.Evol.30,1720-1728(2013).19Posada,D.&Buckley,T.R.Modelselectionandmodelaveraginginphylogenetics:AdvantagesofakaikeinformationcriterionandBayesianapproachesoverlikelihoodratiotests.Syst.Biol.53,793-808(2004).20Darriba,D.,Taboada,G.L.,Doallo,R.&Posada,D.jModelTest2:moremodels,newheuristicsand245parallelcomputing.Nat.Methods9,772-772(2012).21Stamatakis,A.RAxML-VI-HPC:maximumlikelihood-basedphylogeneticanalyseswiththousandsoftaxaandmixedmodels.Bioinformatics22,2688-2690(2006).22Huelsenbeck,J.P.&Ronquist,F.MRBAYES:Bayesianinferenceofphylogenetictrees.Bioinformatics17,754-755(2001).25023Wang,K.&Zhao,H.Birdsgenerallycarryasmallrepertoireofbittertastereceptorgenes.GenomeBiolEvol7,2705-2715(2015).24Lu,Q.,Wang,K.,Lei,F.,Yu,D.&Zhao,H.Penguinsreducedolfactoryreceptorgenescommontootherwaterbirds.ScientificReports6,31671(2016).25Liu,L.,Yu,L.&Edwards,S.V.Amaximumpseudo-likelihoodapproachforestimatingspeciestrees-7-
中国科技论文在线http://www.paper.edu.cn255underthecoalescentmodel.BMCEvol.Biol.10,302(2010).26Abascal,F.,Zardoya,R.&Posada,D.ProtTest:selectionofbest-fitmodelsofproteinevolution.Bioinformatics21,2104-2105(2005).27Vila,M.&Bjorklund,M.Theutilityoftheneglectedmitochondrialcontrolregionforevolutionarystudiesinlepidoptera(Insecta).J.Mol.Evol.58,280-290(2004).26028Boore,J.L.Animalmitochondrialgenomes.NucleicAcidsRes.27,1767-1780(1999).29Pollard,D.A.,Iyer,V.N.,Moses,A.M.&Eisen,M.B.WidespreaddiscordanceofgenetreeswithspeciestreeinDrosophila:evidenceforincompletelineagesorting.PLoSGenet.2,e173(2006).265三种萤火虫的线粒体基因组测序和18种甲虫的系统发育分析汪凯,洪炜,焦恒武,穆凤娟,赵华斌(武汉大学生命科学学院,武汉430072)270摘要:为了验证基于二代测序获得的转录组数据的系统发育推断,利用传统的一代测序获得了黄缘萤(Aquaticficta)和武汉萤(Aquaticwuhana)的线粒体基因组全序列和云南扁萤(Lamprigerayunnana)的线粒体基因组接近全序列;从4种发光甲虫的转录组序列中查找到所有的线粒体编码基因序列;再从公共数据库中提取了12种近源甲虫的线粒体基因组全序列。由于黄缘萤的线粒体基因组序列同时有一代和二代测序的数据,本研究共分析了18275种甲虫的线粒体基因组序列。对所有甲虫的线粒体编码序列进行对位排列后,利用最大似然法和贝叶斯推断的方法,将18种甲虫的线粒体数据进行系统发育重建。结果显示,雌光萤与萤科的萤火虫具有显著不同的系统位置,支持将雌光萤独立成一个科,与之前的转录组分析结果一致。因此,该研究不仅提供了新的线粒体全序列数据,将为未来的系统发育分析提供数据,同时也指出了使用线粒体基因和核基因进行综合分析对系统发育研究的重280要性。关键词:动物学;萤火虫;系统发育;线粒体基因组;线粒体中图分类号:Q951+.3-8-'
您可能关注的文档
- MicroRNAs对间充质干细胞成骨向分化的调控作用.pdf
- MicroRNA在其他类型细胞转分化为神经元中的作用.pdf
- miR-590-3p及TFAM在氡致肺损伤中的表达改变.pdf
- R1234yf燃烧产物HF的实验研究.pdf
- St. Jude 医院儿童急性淋巴细胞白血病系列研究方案的发展.pdf
- TGR5通过抑制S1PS1P2通路、抵抗高糖诱导的肾小球系膜细胞炎症纤维化.pdf
- “三法三穴”不同刺激参数对坐骨神经损伤大鼠痛温觉功能的影响.pdf
- 一种多级孔生物活性玻璃的制备及固载溶菌酶研究.pdf
- 一种应对云环境下在线聚集的数据倾斜方法.pdf
- 三维颈动脉超声图像中斑块的纹理特征分析.pdf
- 不同Wx等位组合对杂交稻天优3611品质的影响.pdf
- 不同分辨率DEM与坡度关系分析.pdf
- 不同强度与时间高压静电辐射处理对麦长管蚜生长发育与繁殖的影响.pdf
- 个性特征对洪灾后创伤后应激障碍慢性化的影响.pdf
- 临夏学龄少数民族儿童患龋情况及相关因素调查分析.pdf
- 从“湿热瘀结”论治子宫内膜异位症.pdf
- 优化新生脉散方对心衰大鼠心功能及NT-proBNP水平影响的实验研究简报.pdf
- 光敏感型水凝胶的研究进展.pdf
相关文档
- 施工规范CECS140-2002给水排水工程埋地管芯缠丝预应力混凝土管和预应力钢筒混凝土管管道结构设计规程
- 施工规范CECS141-2002给水排水工程埋地钢管管道结构设计规程
- 施工规范CECS142-2002给水排水工程埋地铸铁管管道结构设计规程
- 施工规范CECS143-2002给水排水工程埋地预制混凝土圆形管管道结构设计规程
- 施工规范CECS145-2002给水排水工程埋地矩形管管道结构设计规程
- 施工规范CECS190-2005给水排水工程埋地玻璃纤维增强塑料夹砂管管道结构设计规程
- cecs 140:2002 给水排水工程埋地管芯缠丝预应力混凝土管和预应力钢筒混凝土管管道结构设计规程(含条文说明)
- cecs 141:2002 给水排水工程埋地钢管管道结构设计规程 条文说明
- cecs 140:2002 给水排水工程埋地管芯缠丝预应力混凝土管和预应力钢筒混凝土管管道结构设计规程 条文说明
- cecs 142:2002 给水排水工程埋地铸铁管管道结构设计规程 条文说明